Welcome to Mendelian Randomization Analysis
MRanalysis is an interactive R Shiny application designed for Mendelian randomization analysis.
Introduction
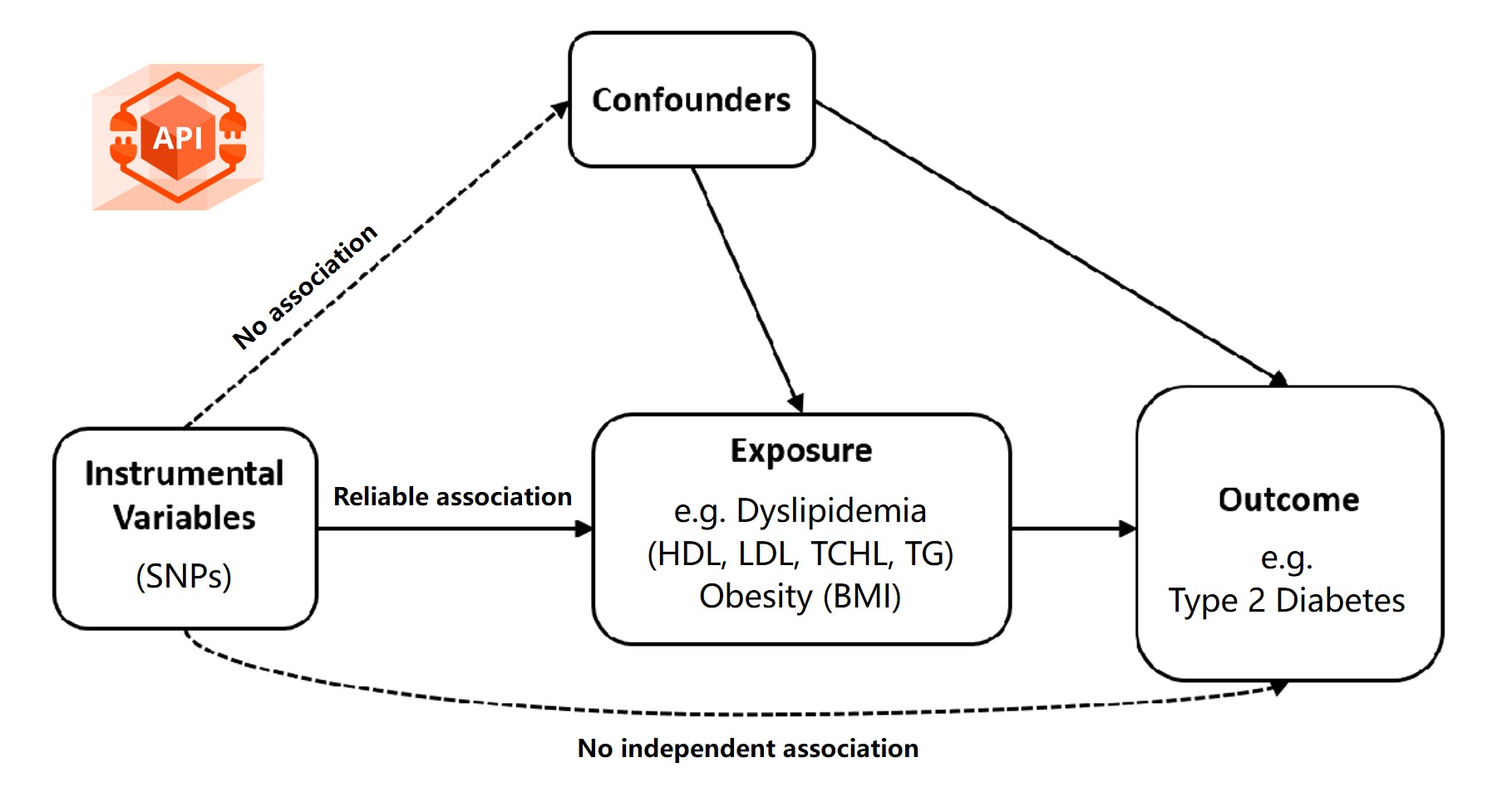
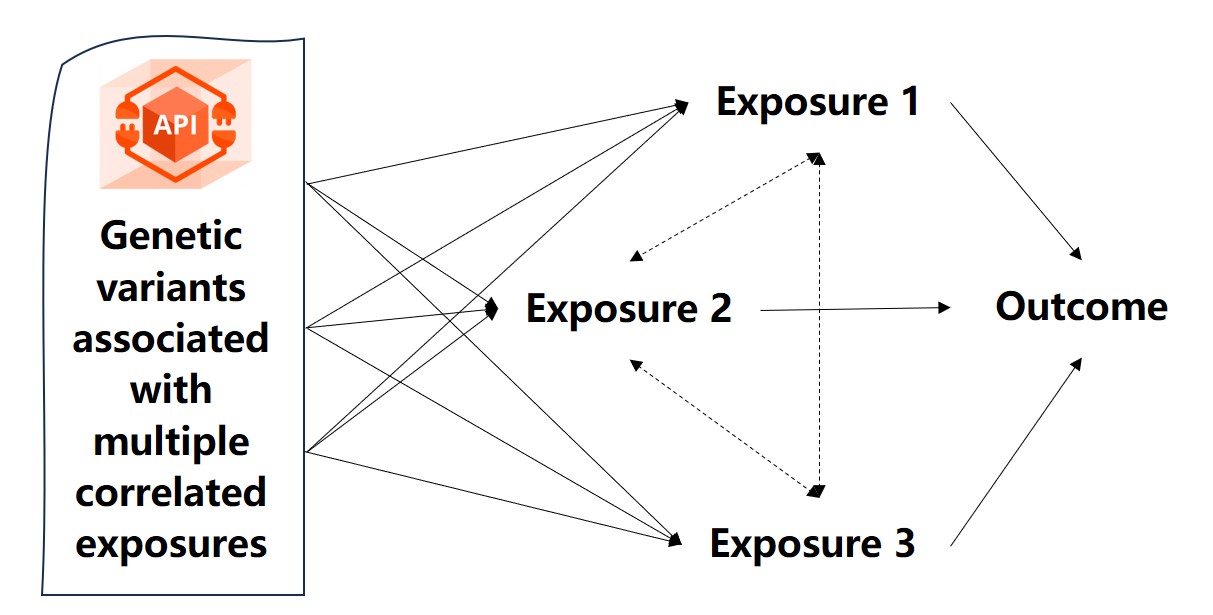
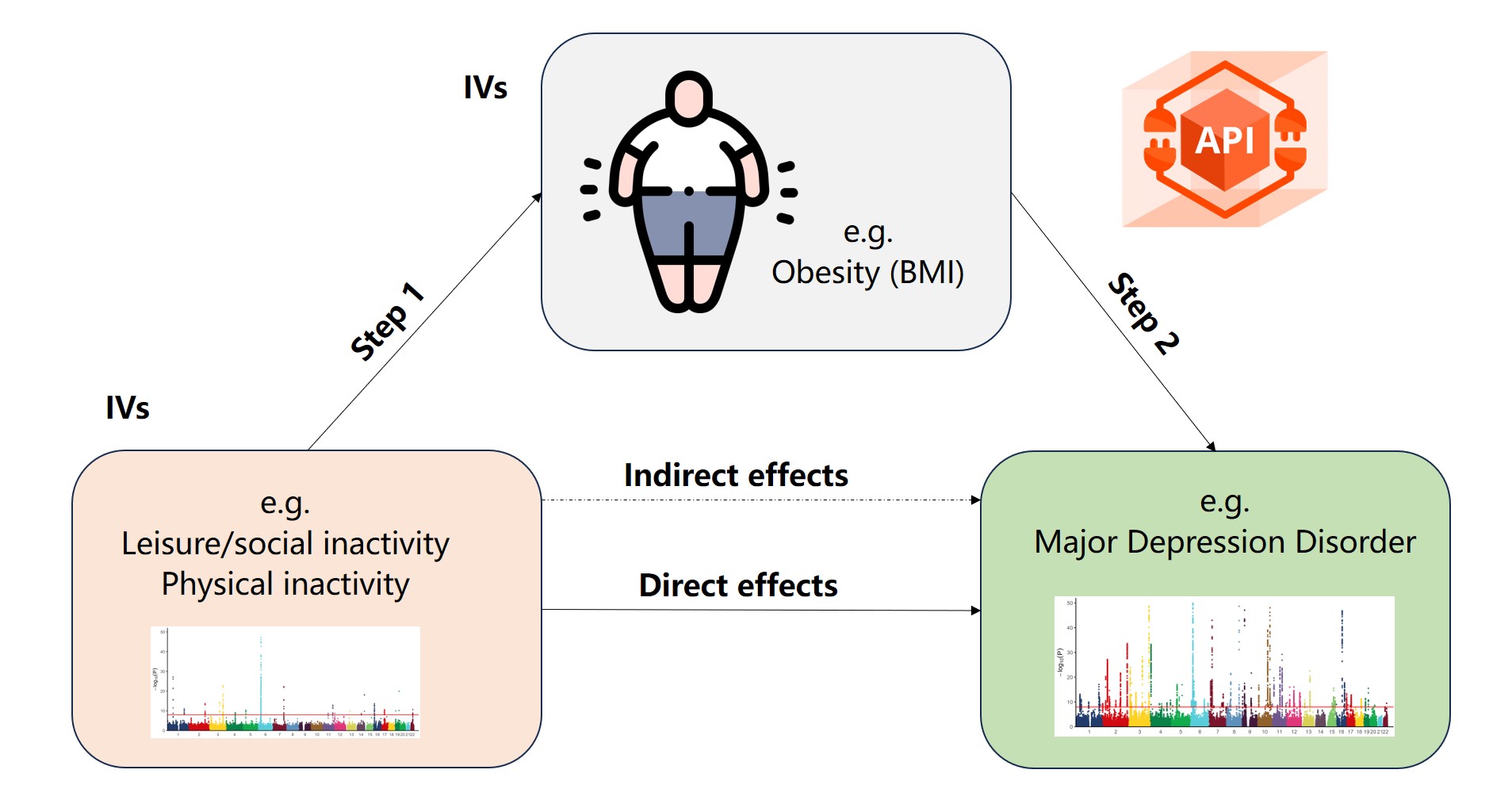
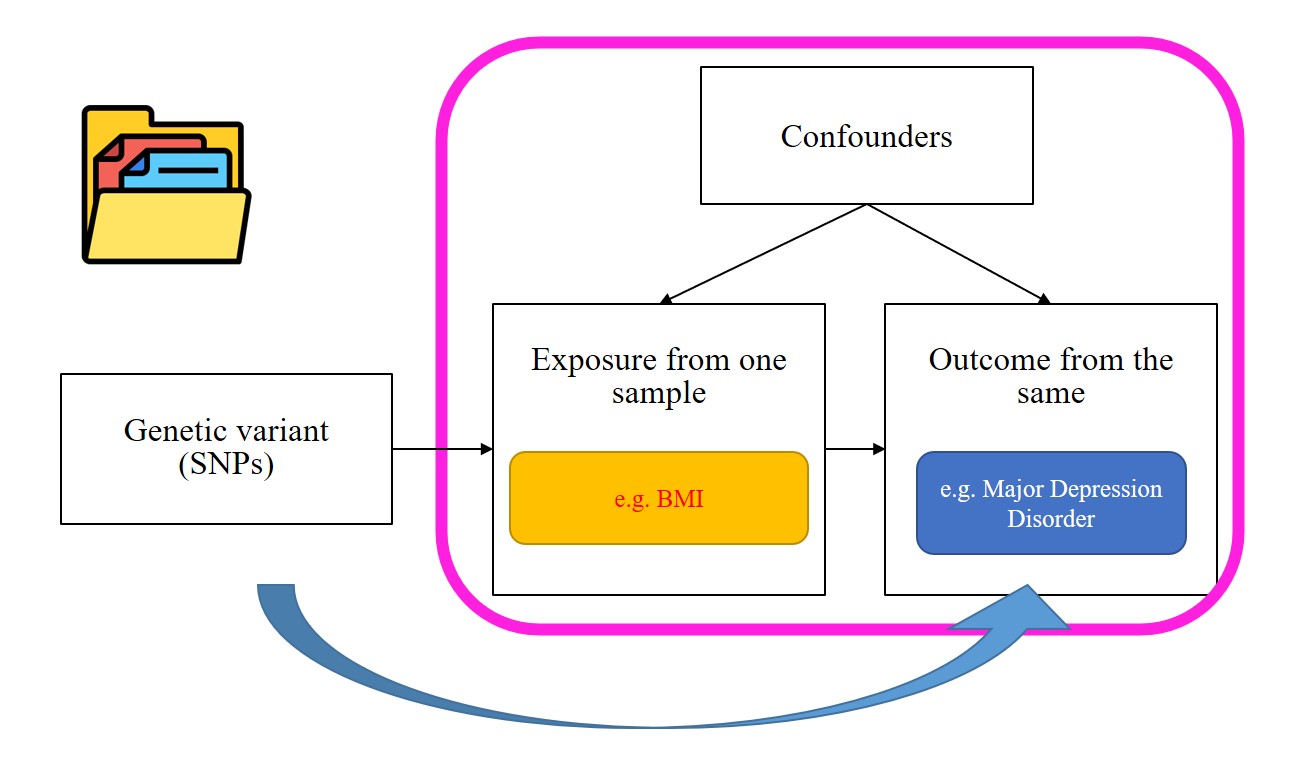
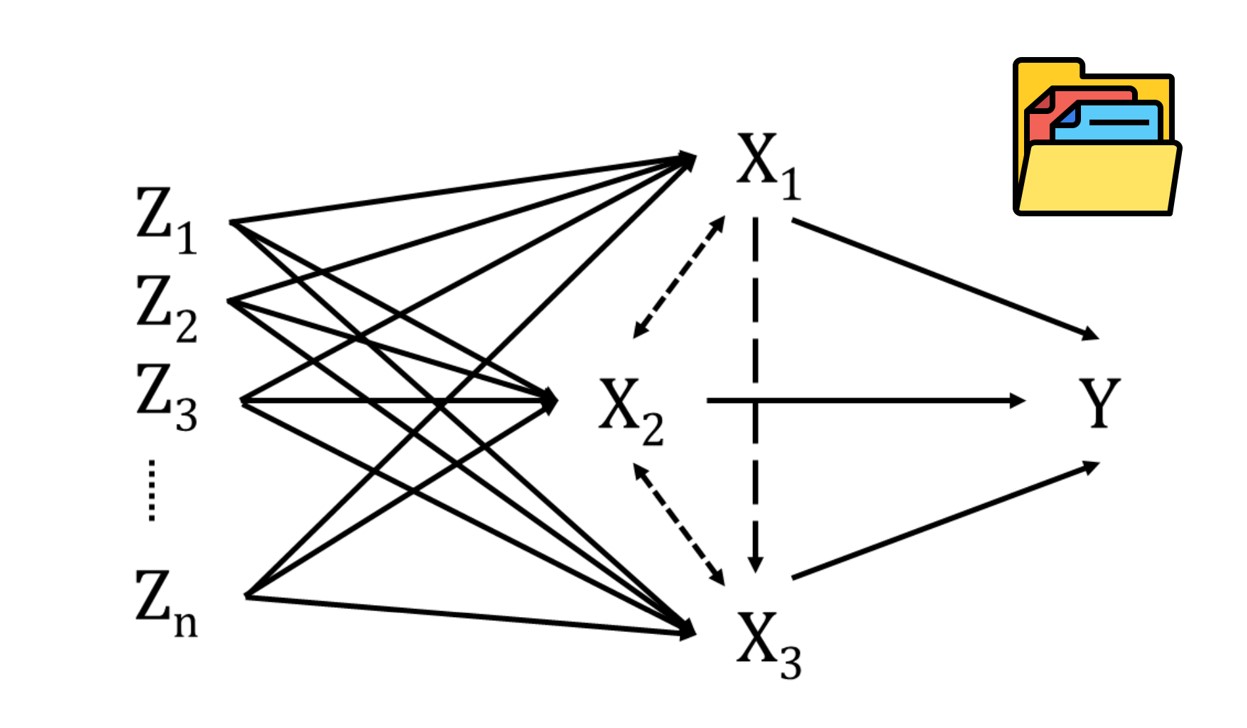
Mendelian randomization (MR) has emerged as a powerful epidemiological method for inferring causal relationships between exposures and outcomes using genome-wide association study (GWAS) summary data. By leveraging instrumental variables (IVs), such as single nucleotide polymorphisms (SNPs), MR can revolutionize our understanding of disease etiology, inform public health strategies, and accelerate drug discovery.
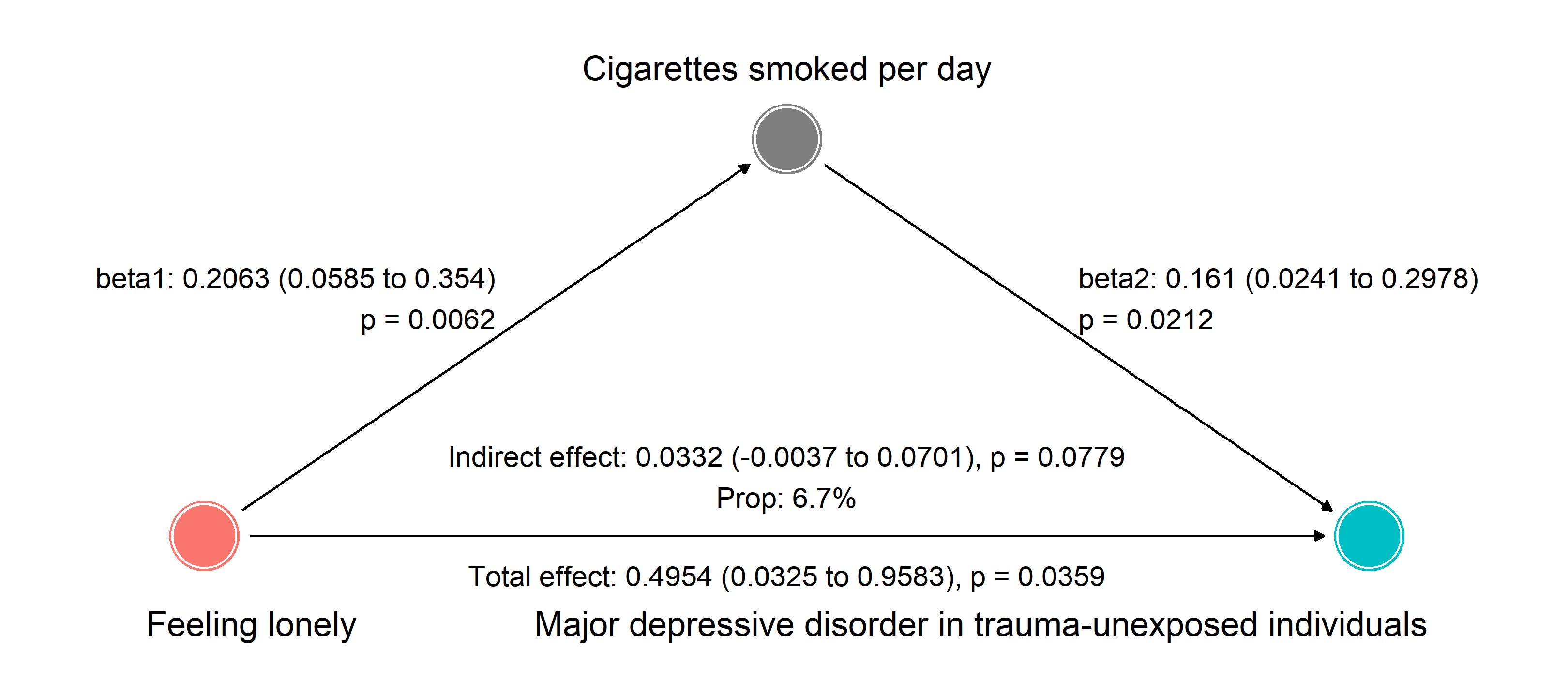
However, the widespread adoption of MR is hindered by several challenges, including inconsistent GWAS data formats, lack of standardized workflows, the need for extensive programming expertise, and limitations in data visualization and interpretability. To address these challenges, we introduce MRanalysis , a comprehensive and user-friendly, web-based platform that provides the first integrated and standardized MR analysis workflow. This includes GWAS data quality assessment, power/ sample size estimation, MR analysis, SNP-to-gene enrichment analysis, and data visualization. Built using the R shiny framework, MRanalysis enables users to conduct common MR methods, including univariable, multivariable, and mediation MR analyses through an intuitive, no-code interface .

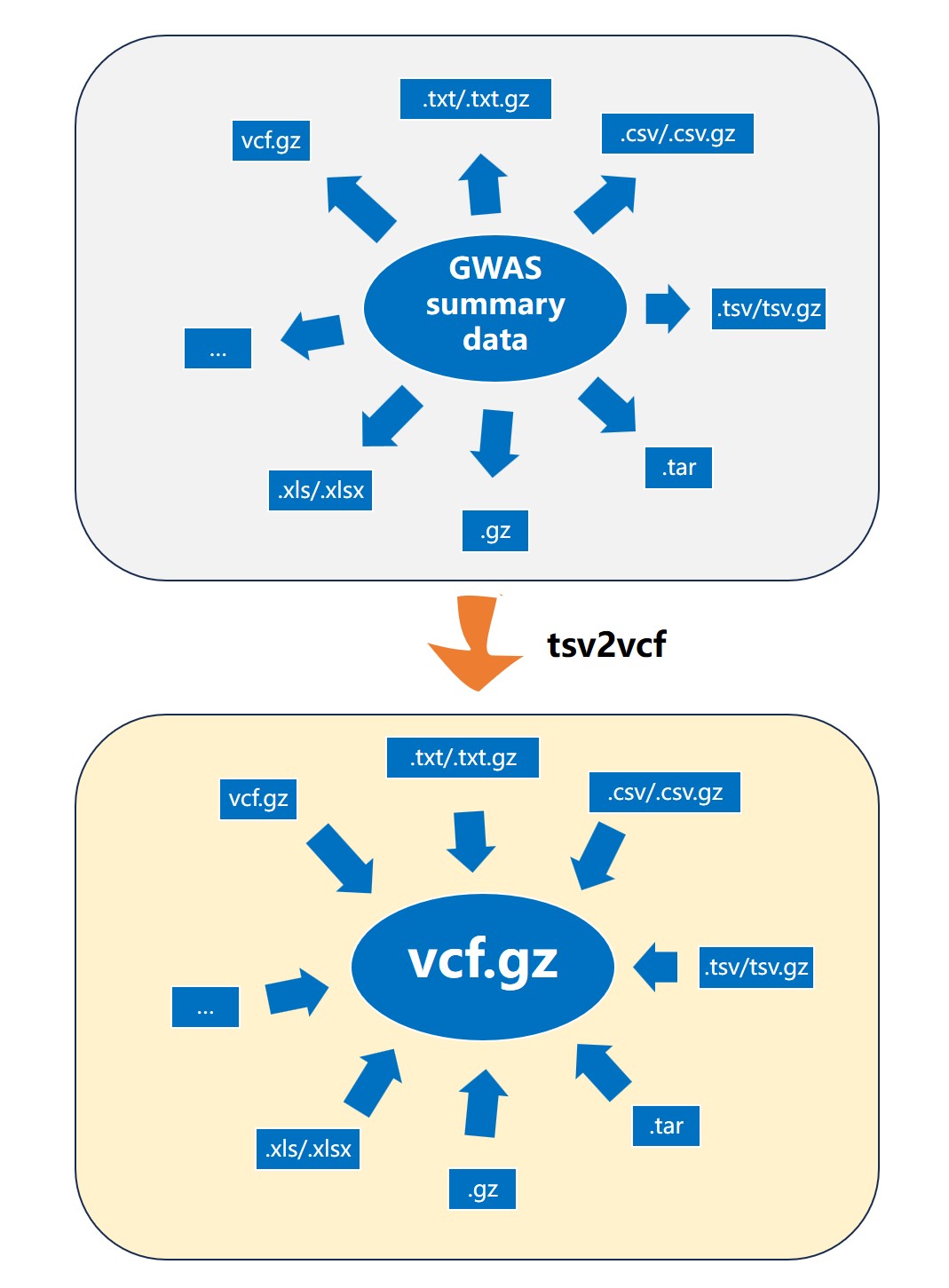
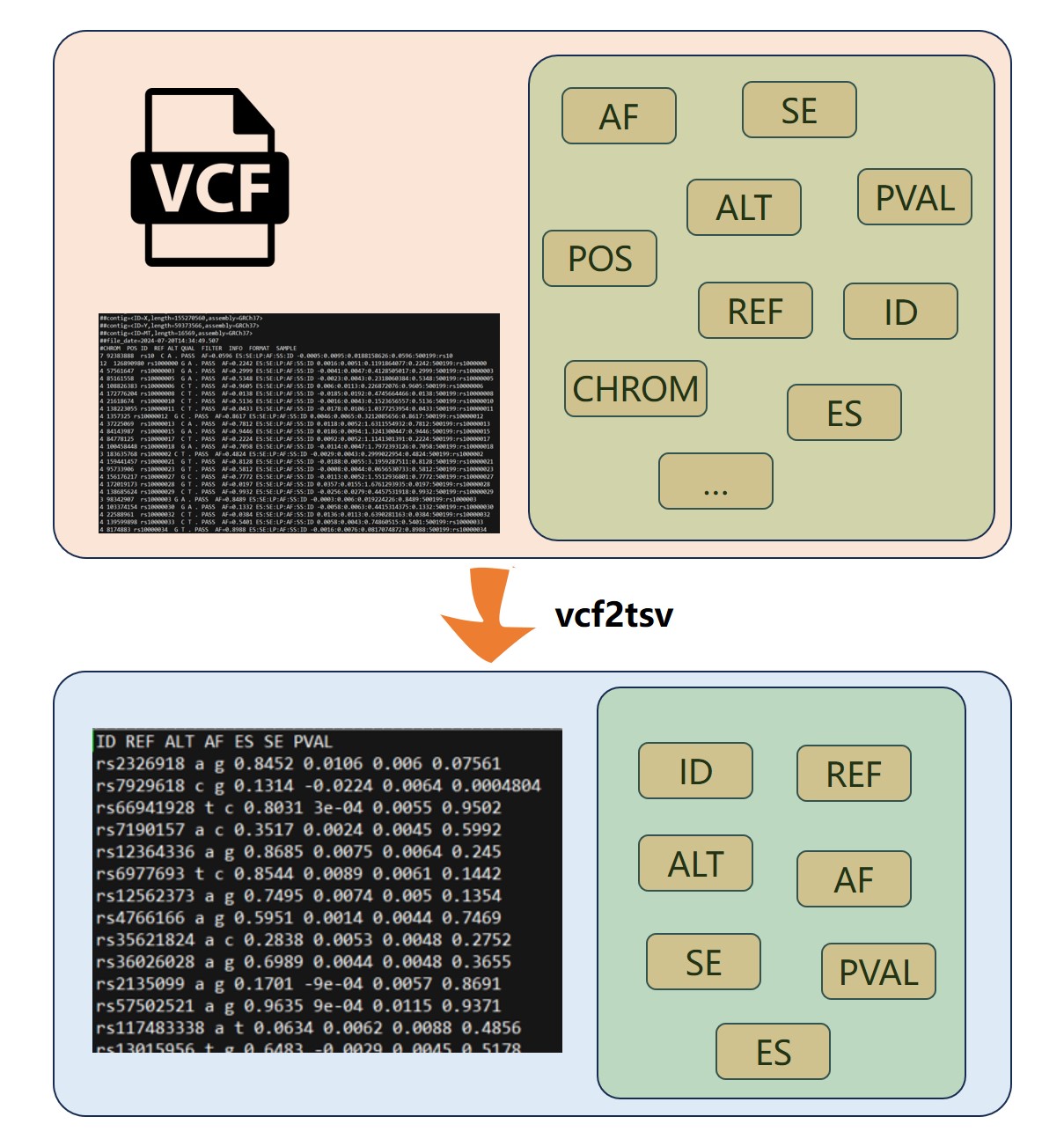
Besides MRanalysis, we developed GWASkit , a standalone, and installation-free tool facilitating rapid GWAS dataset preprocessing before MR analyses, including rs ID conversion, format standardization, and data extraction, with significantly lower conversion time and dramatically higher rs ID conversion accuracy than the current tools. Case studies demonstrate the utility, efficiency, and ease of use of our developed platform and GWASkit tool in real-world scenarios. By lowering barriers to investigating causal genetic relationships, our platform represents a significant advance in making MR more accessible, reliable, and efficient. The increased adoption of MR, facilitated by MRanalysis and GWASkit, can accelerate discoveries in genetic epidemiology, strengthen evidence-based public health strategies, and guide the development of targeted clinical interventions.
📋 Changelog
Latest updates and changes to MRanalysis
-
Version 2.0.0 - August 15, 2025
- 🚢 Docker image & one-click deployment: Released official Docker image for easy local deployment, enabling users to leverage local computing resources for large-scale analysis.
- 🔐 OpenGWAS JWT validation & update: Added token validity check and manual update module for seamless OpenGWAS API connection.
- ⚙️ Flexible IV significance threshold: Users can now set custom P-value thresholds for instrumental variable selection.
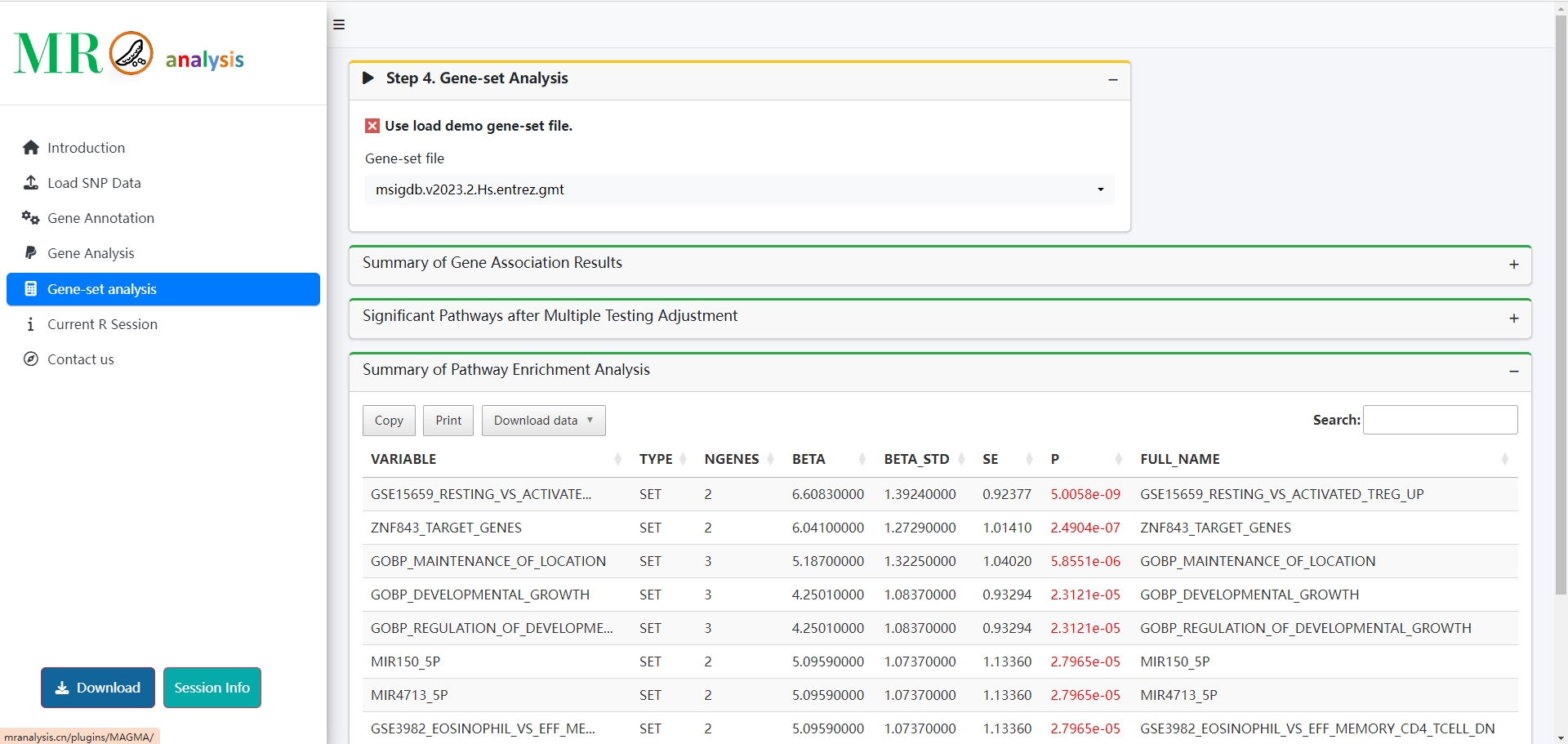
- 🔬 Colocalization analysis: Integrated COLOC.ABF, COLOC.SUSIE, COLOC.PWCoCo, and COLOC.eCAVIAR for robust post-MR validation.
- 🧬 Gene annotation enhanced: Gene symbols and functional/disease annotations are now displayed for easier biological interpretation.
- 🐳 Large-scale GWAS support: Provided Docker image and local deployment guide for handling large datasets; new GWAS format conversion tools added.
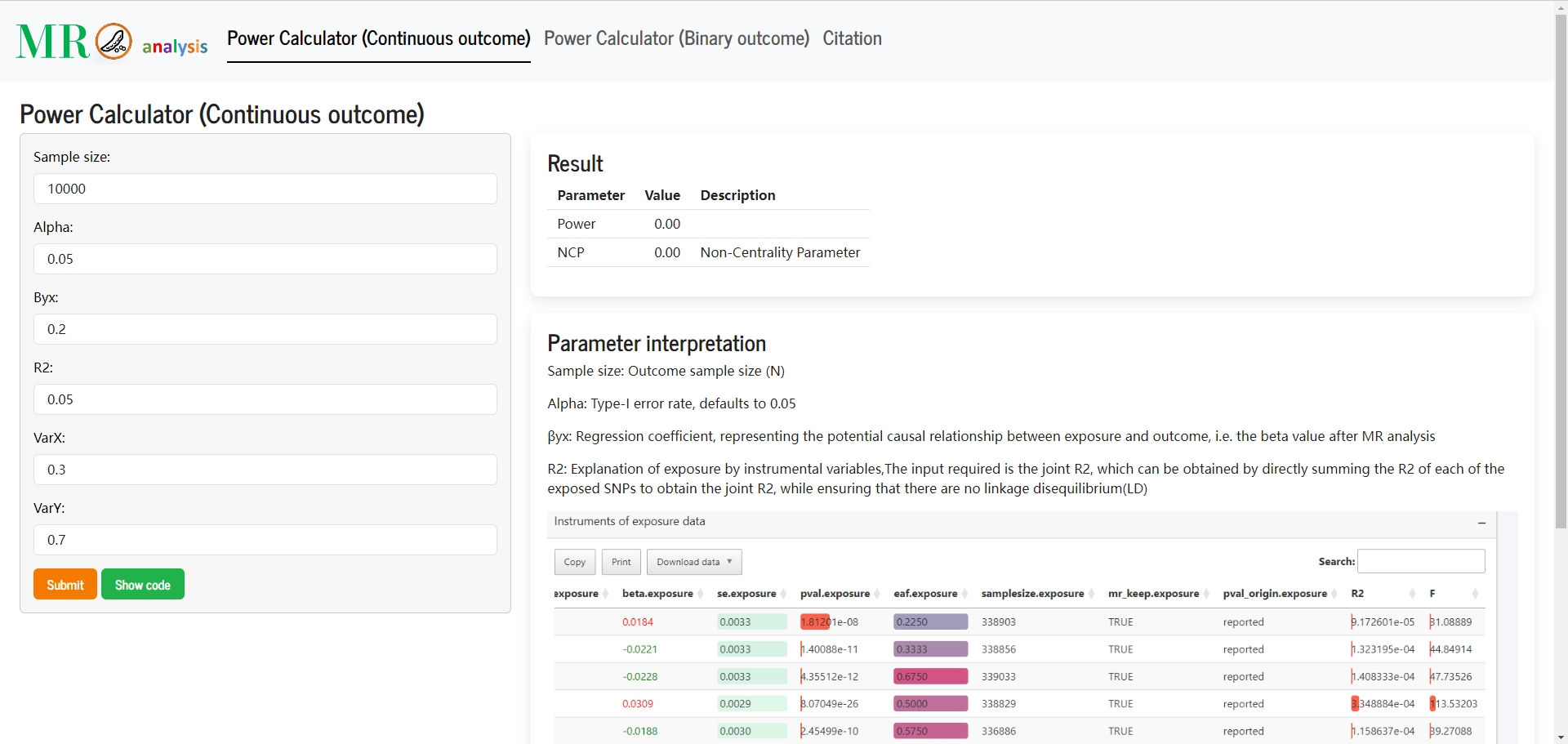
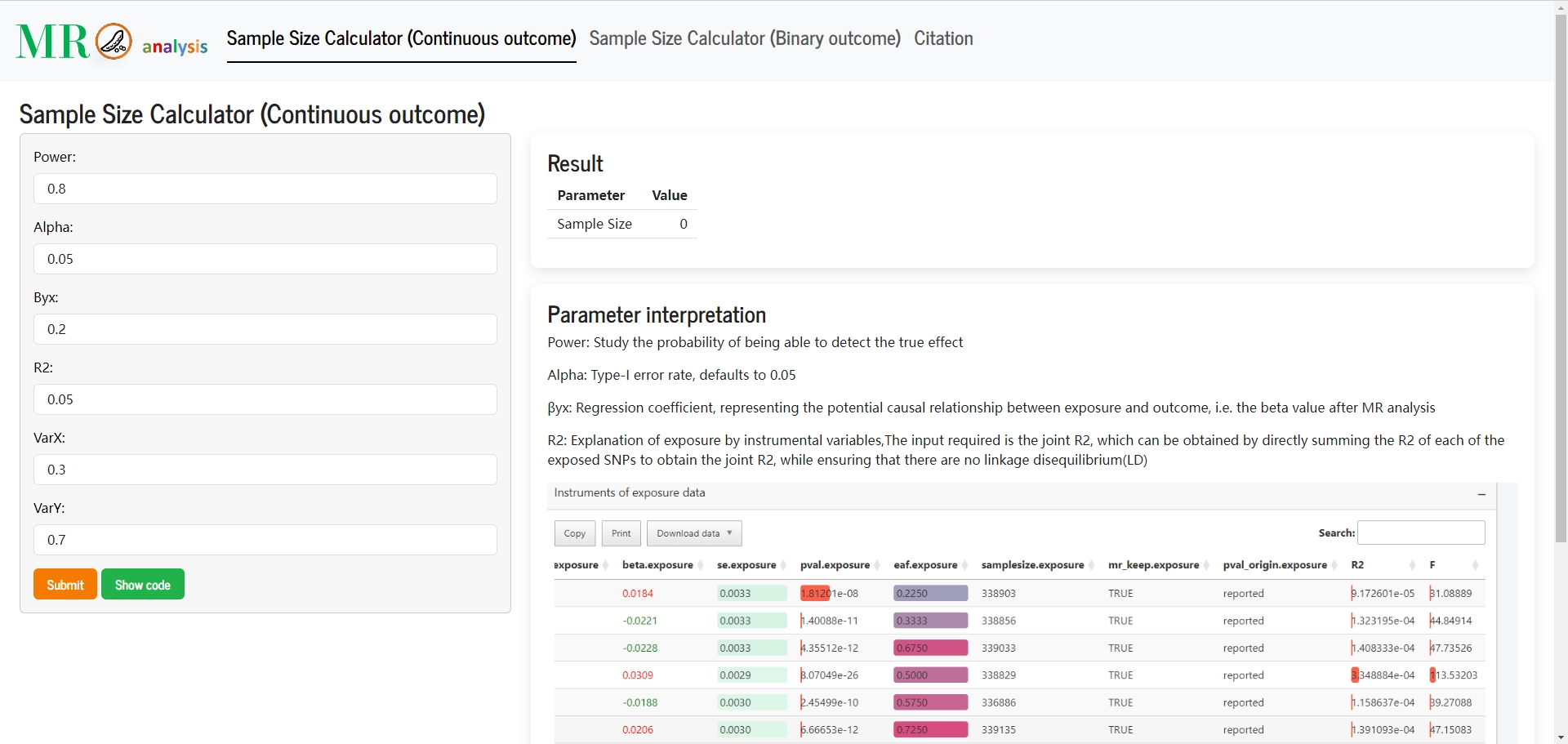
- 📊 Sample size & power calculation: Upgraded calculators with demonstration cases for all three MR analysis types.
- ⚡ Performance improvements: Multivariable MR computation speed increased ~10x; supports local analysis for faster processing.
- 🛡️ Security & accessibility: Upgraded SSL certificate and open-sourced the platform for secure, transparent, and local use.
- 📚 Limitation & interpretation guidance: Added pages on MR limitations, proper use, and interpretation of results, including 2-sample MR caveats.
-
Version 1.8.0 - April 29, 2025
Integrated a lightweight comments widget using Utterances. Feel free to leave any questions or suggestions in the comments section!
-
Version 1.7.0 - April 15, 2025
Added MR-RAPS method for robust Mendelian Randomization analysis.
-
Version 1.6.0 - April 05, 2025
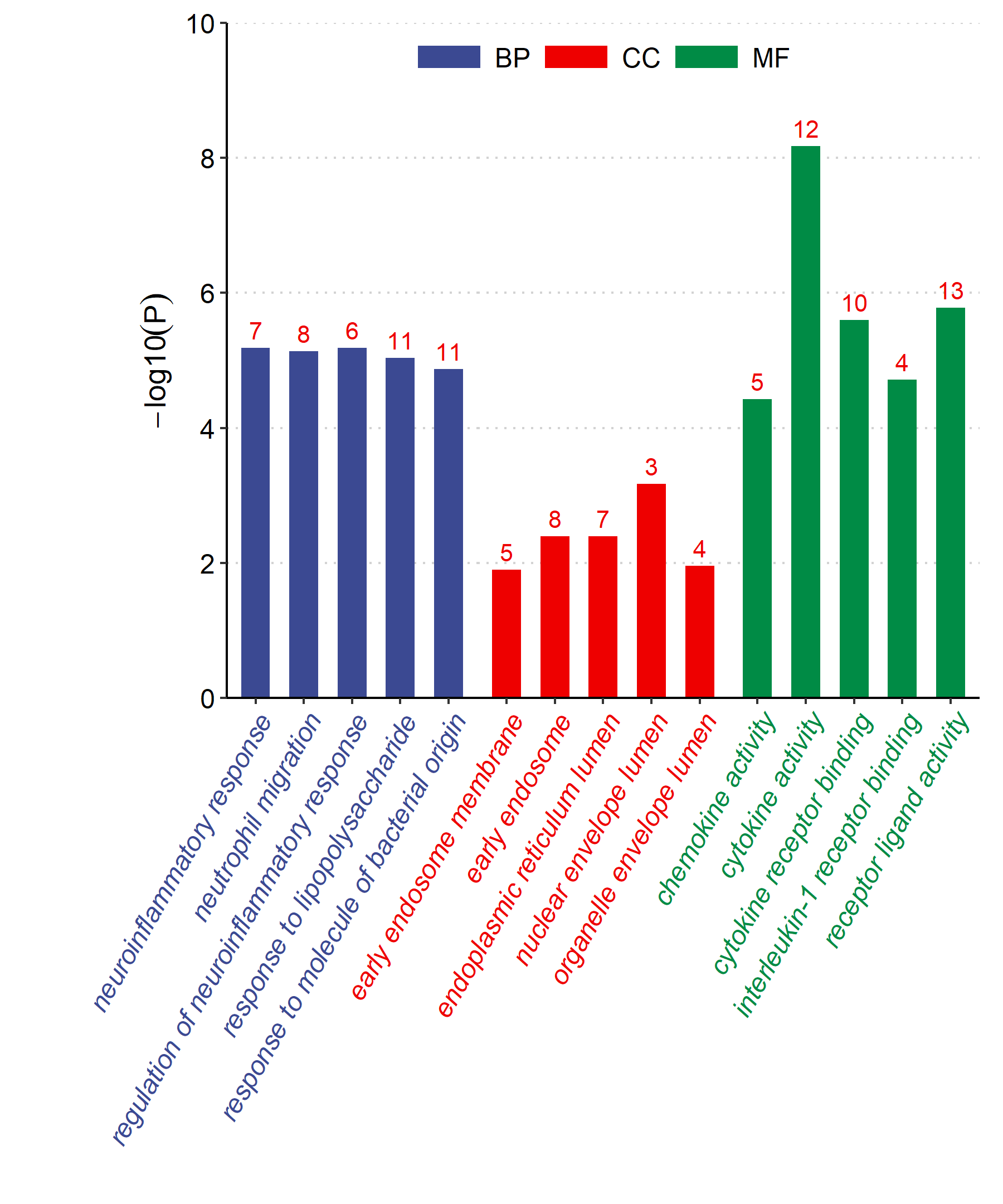
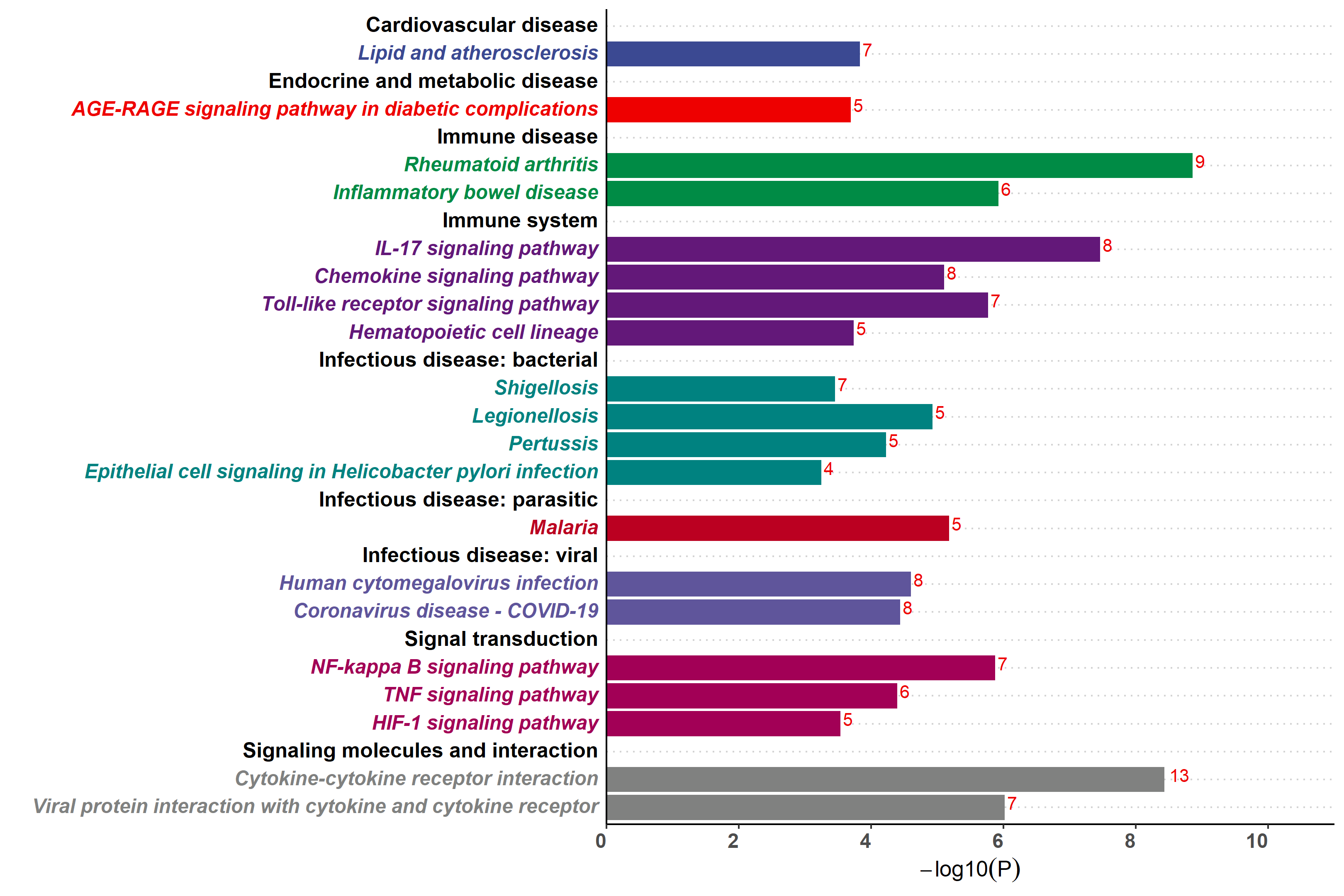
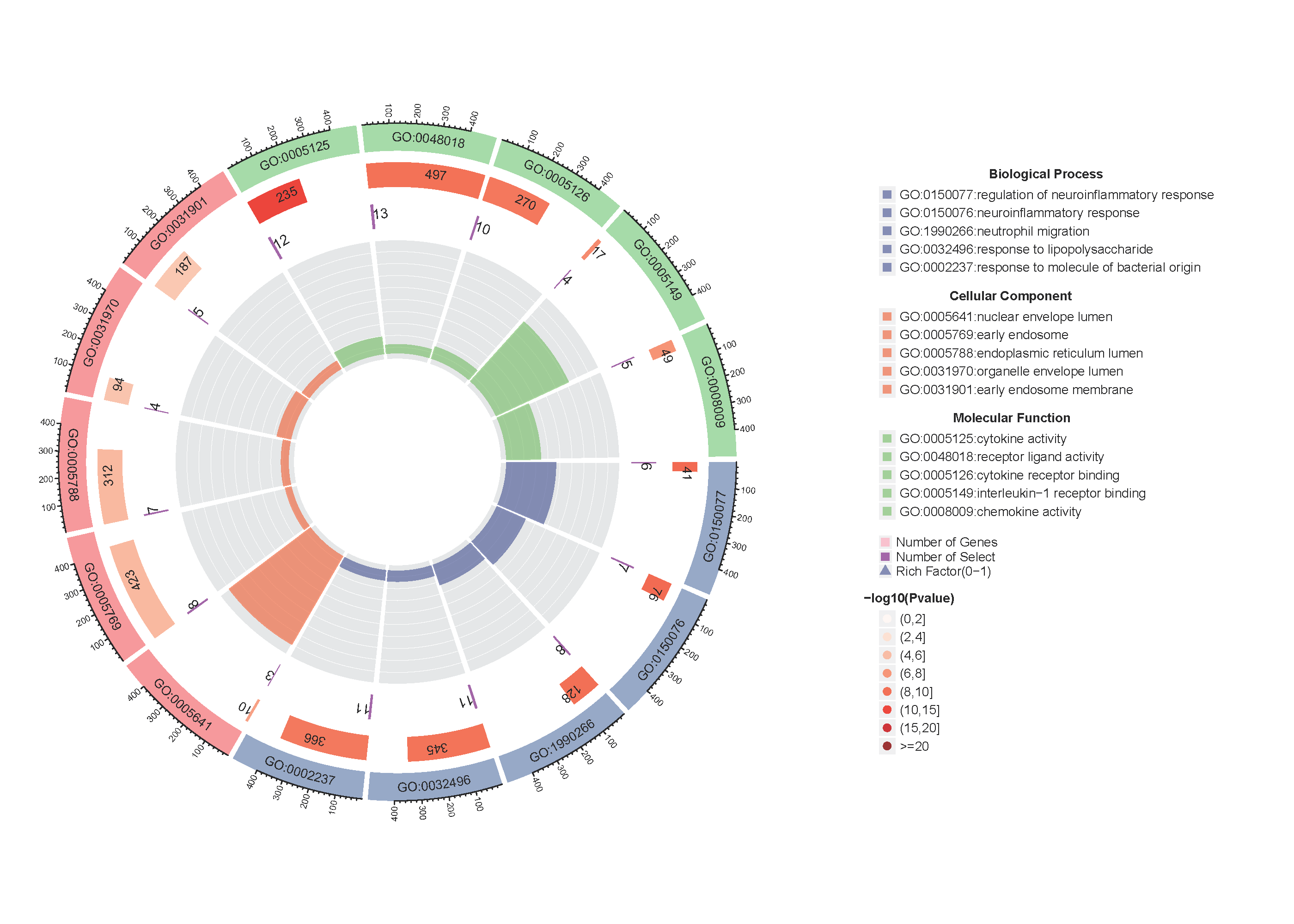
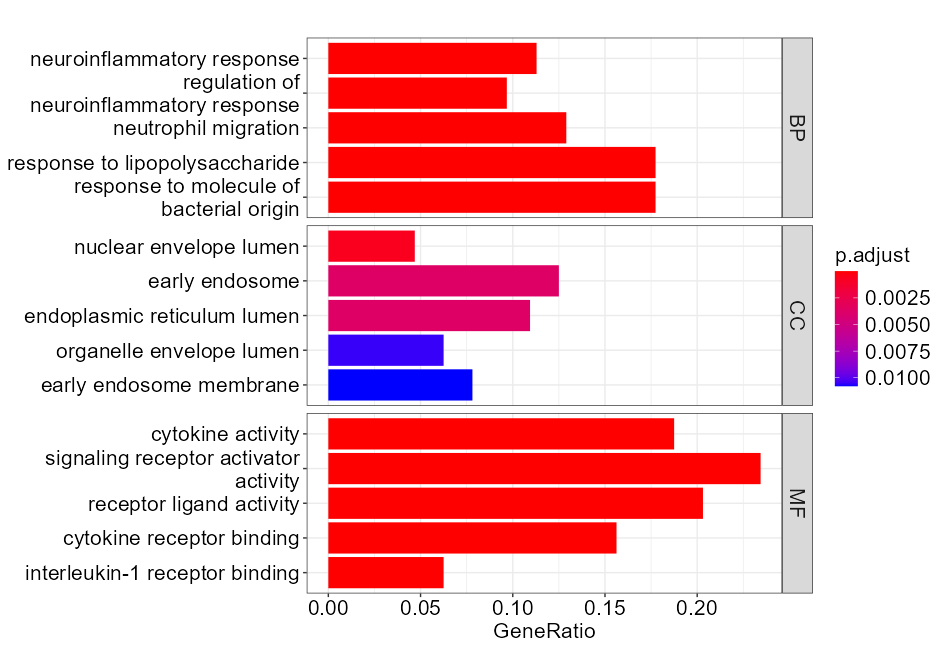
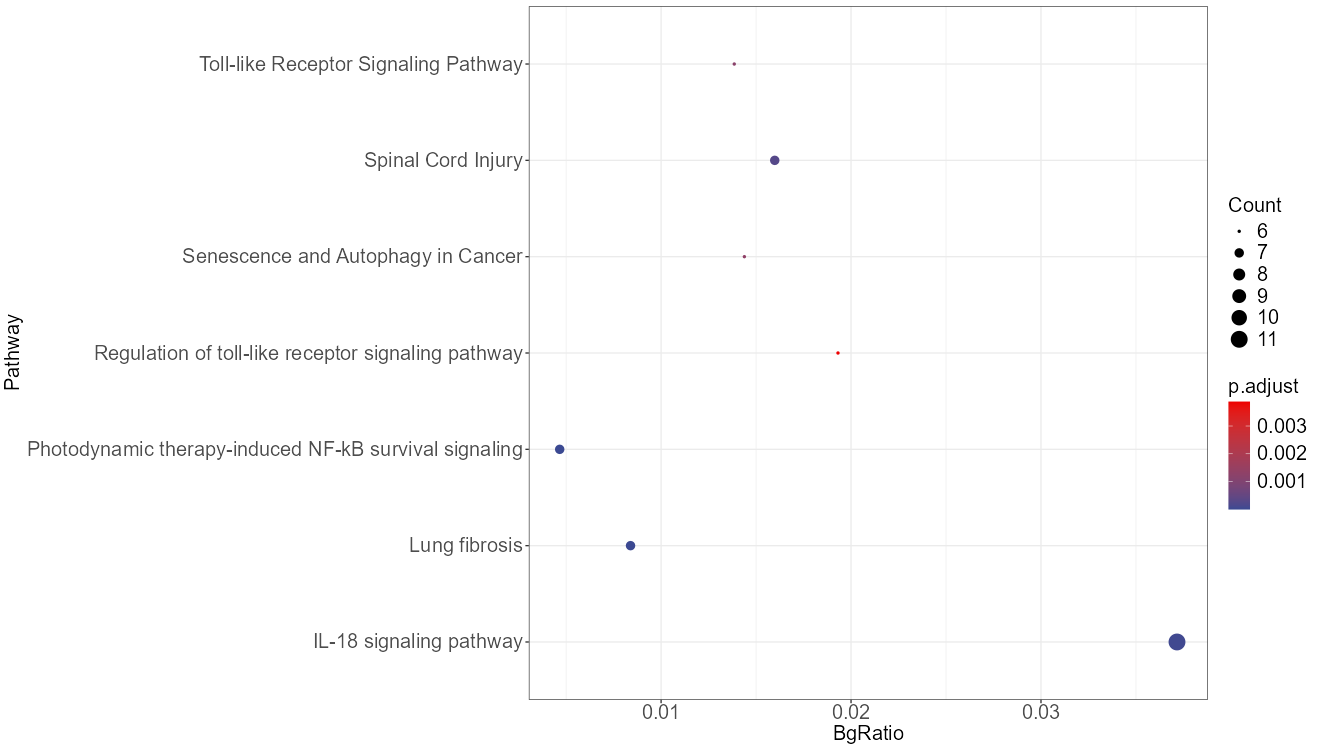
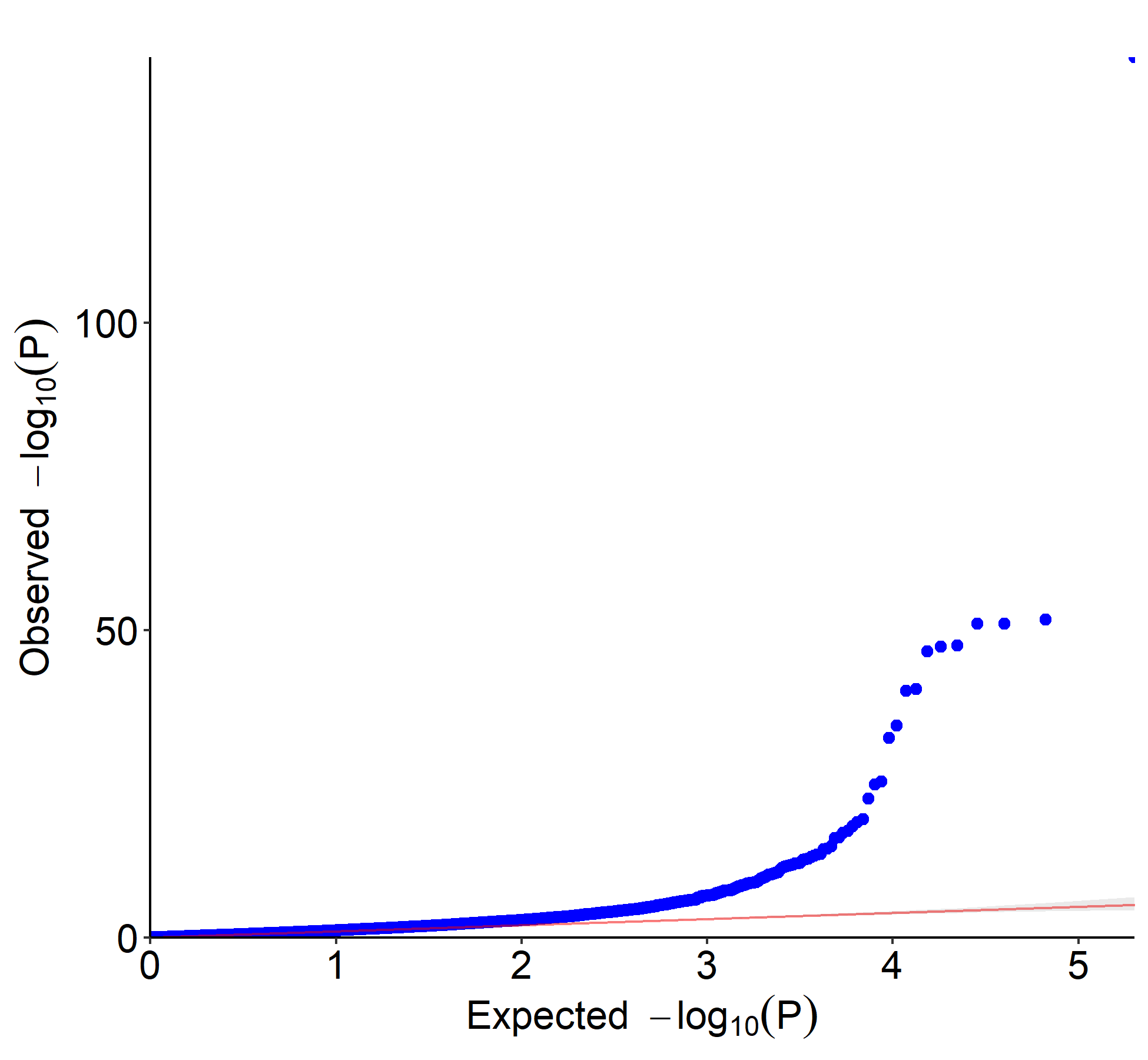
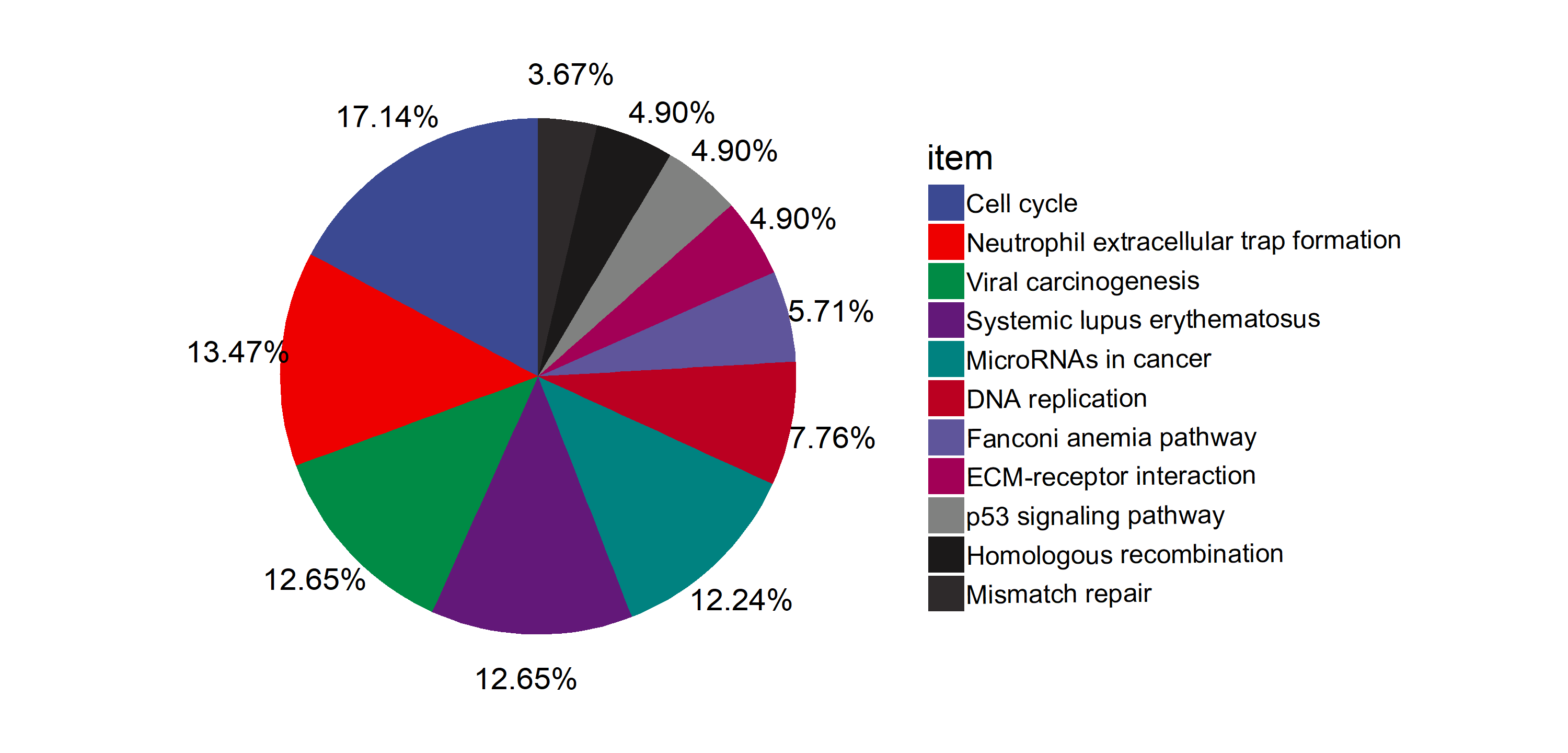
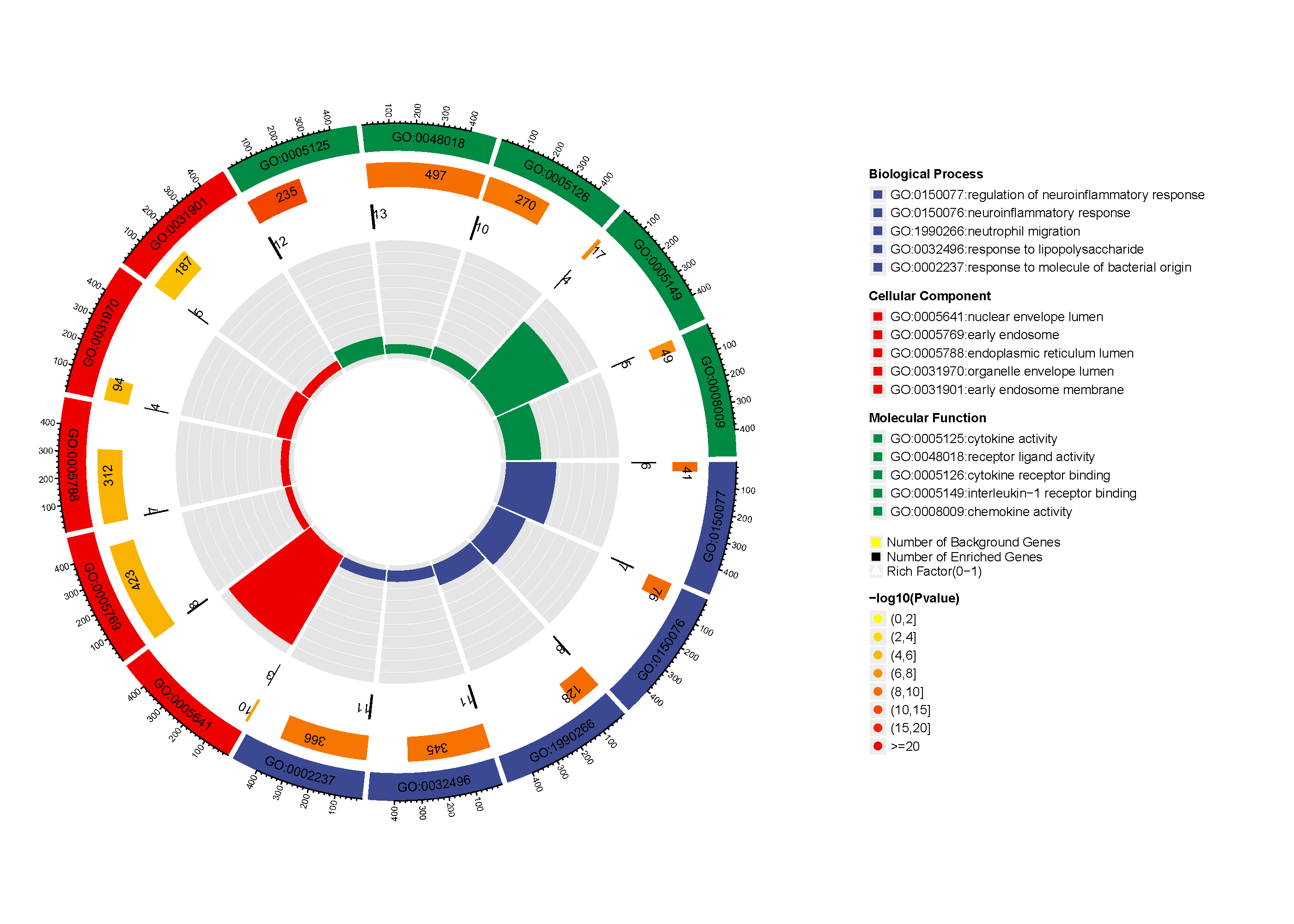
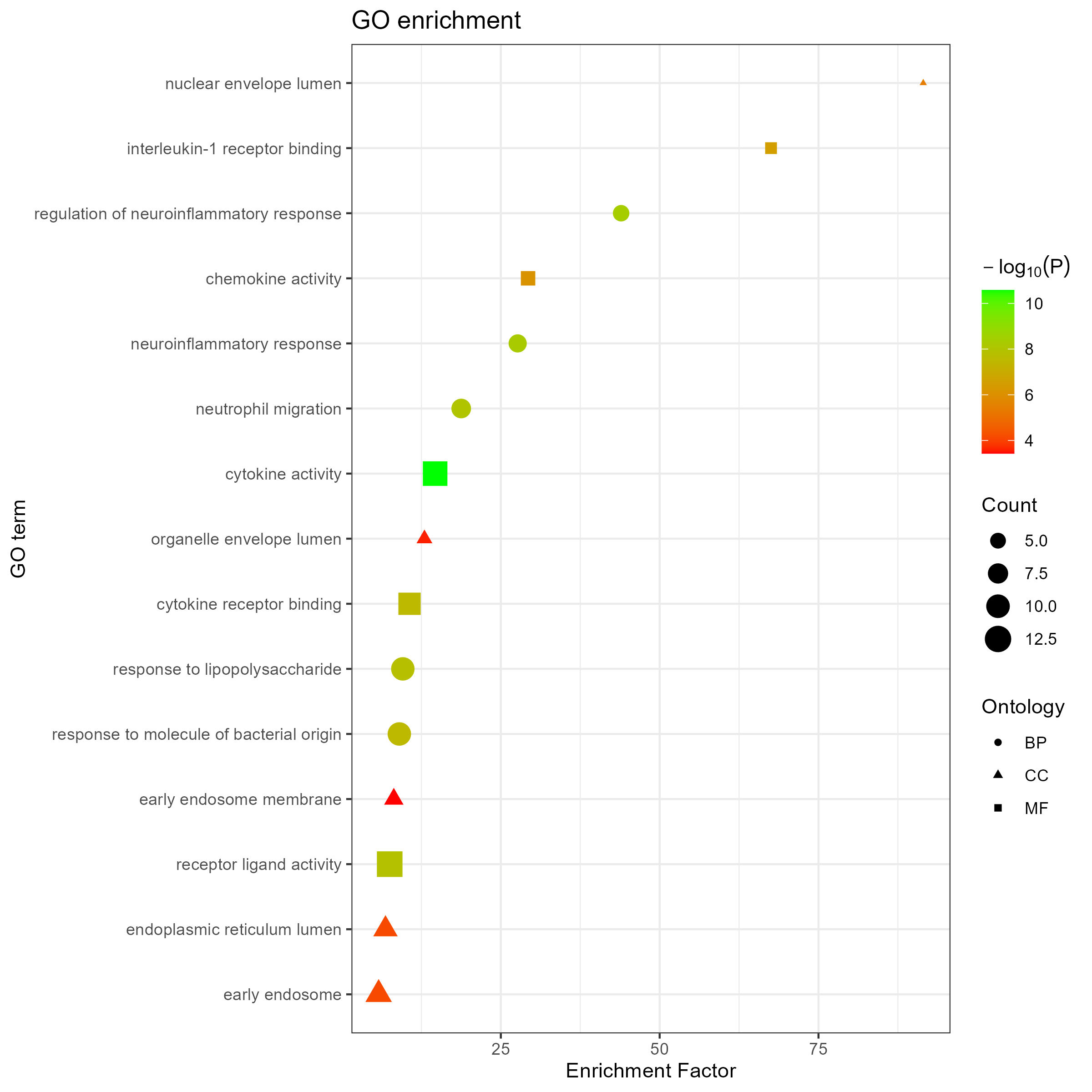
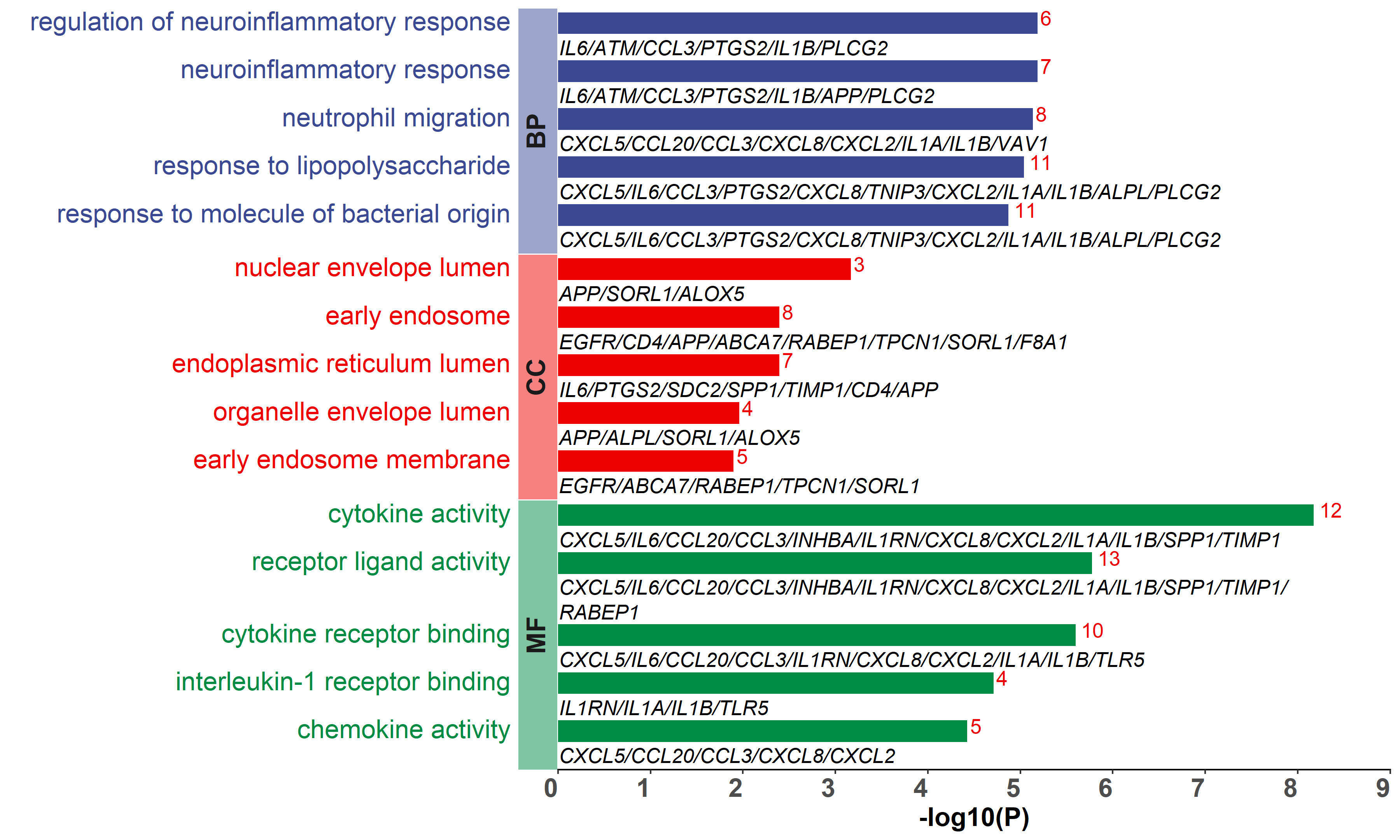
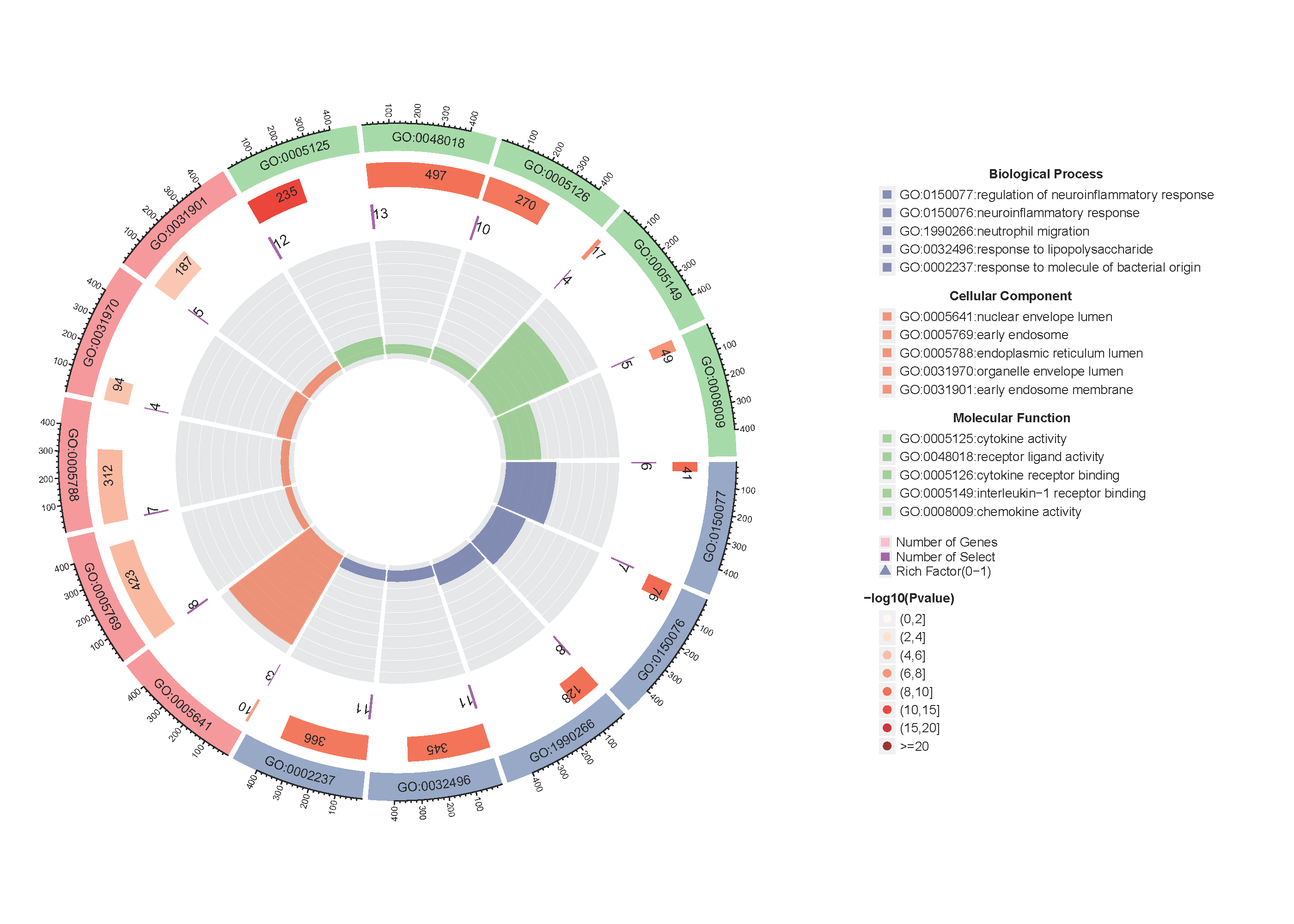
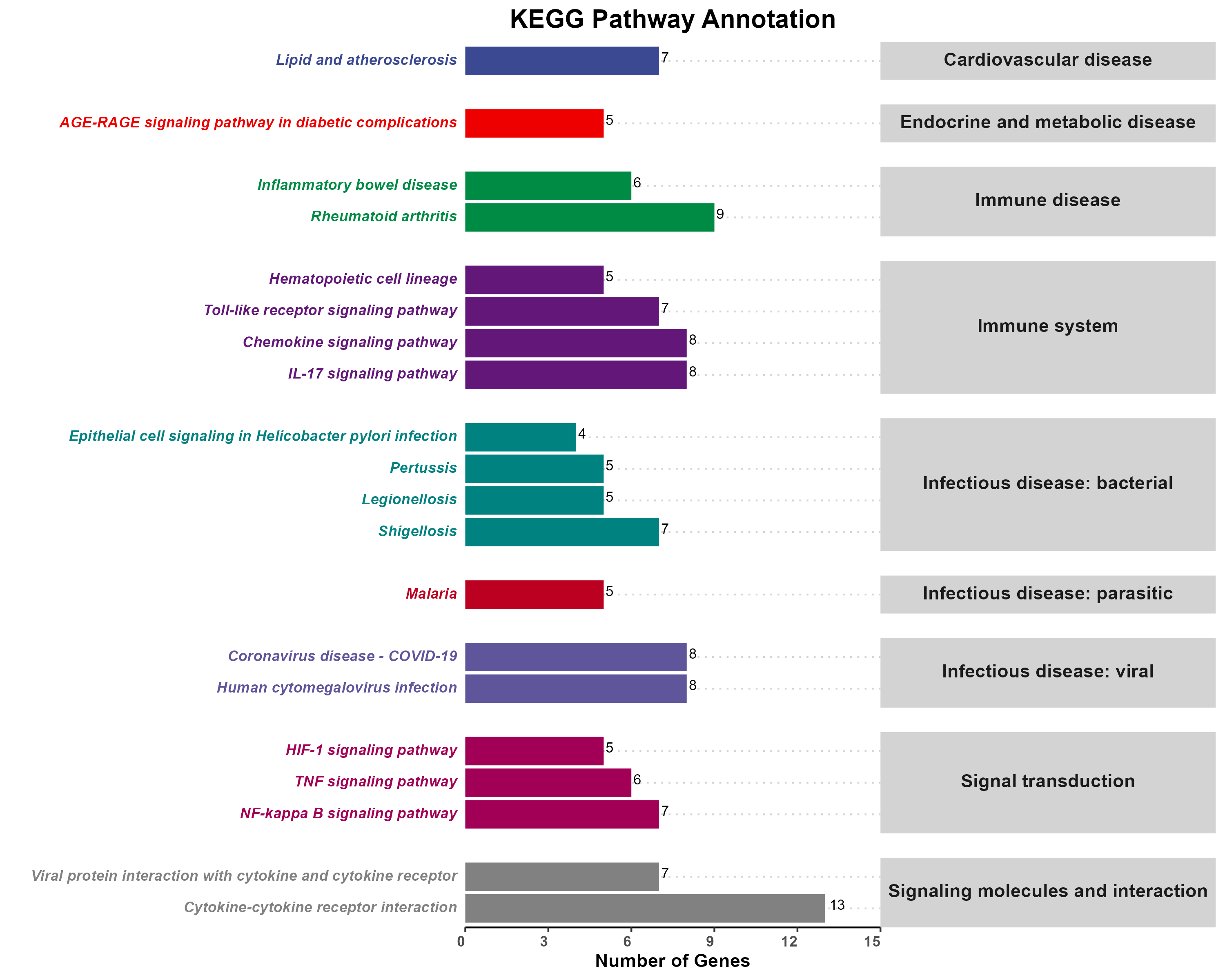
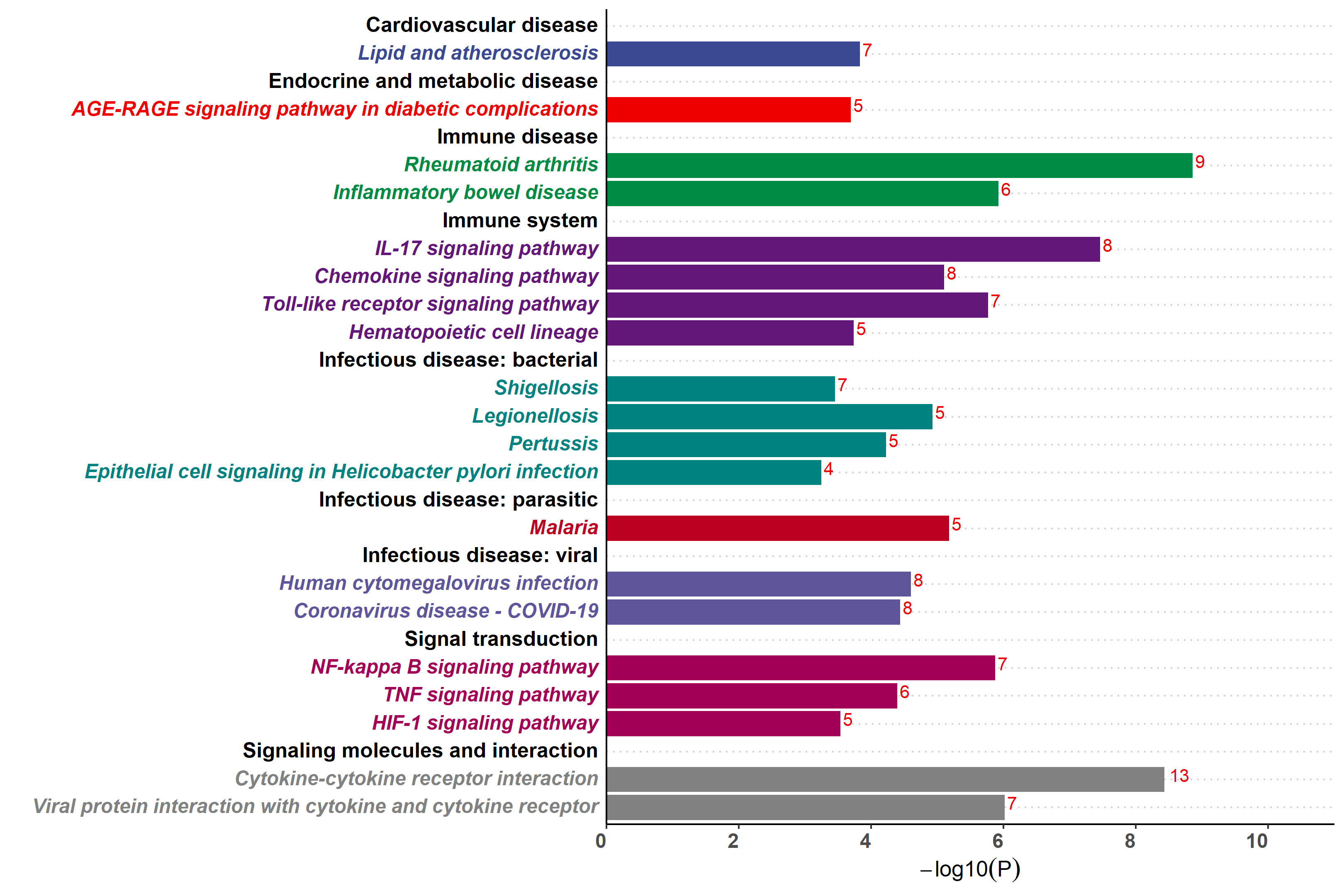
Added support for new visualization tools: Circos Plot (GO) and Bar Plot2 (KEGG).
-
Version 1.5.2 - March 30, 2025
Fixed bug in Manhattan plot rendering for large datasets. Improved performance by 30%.
-
Version 1.5.1 - March 20, 2025
Enhanced data export capabilities with additional file format options and customization settings.
-
Version 1.5.0 - July 26, 2024
Major update to the website's styles.
-
Version 1.4.0 - June 12, 2024
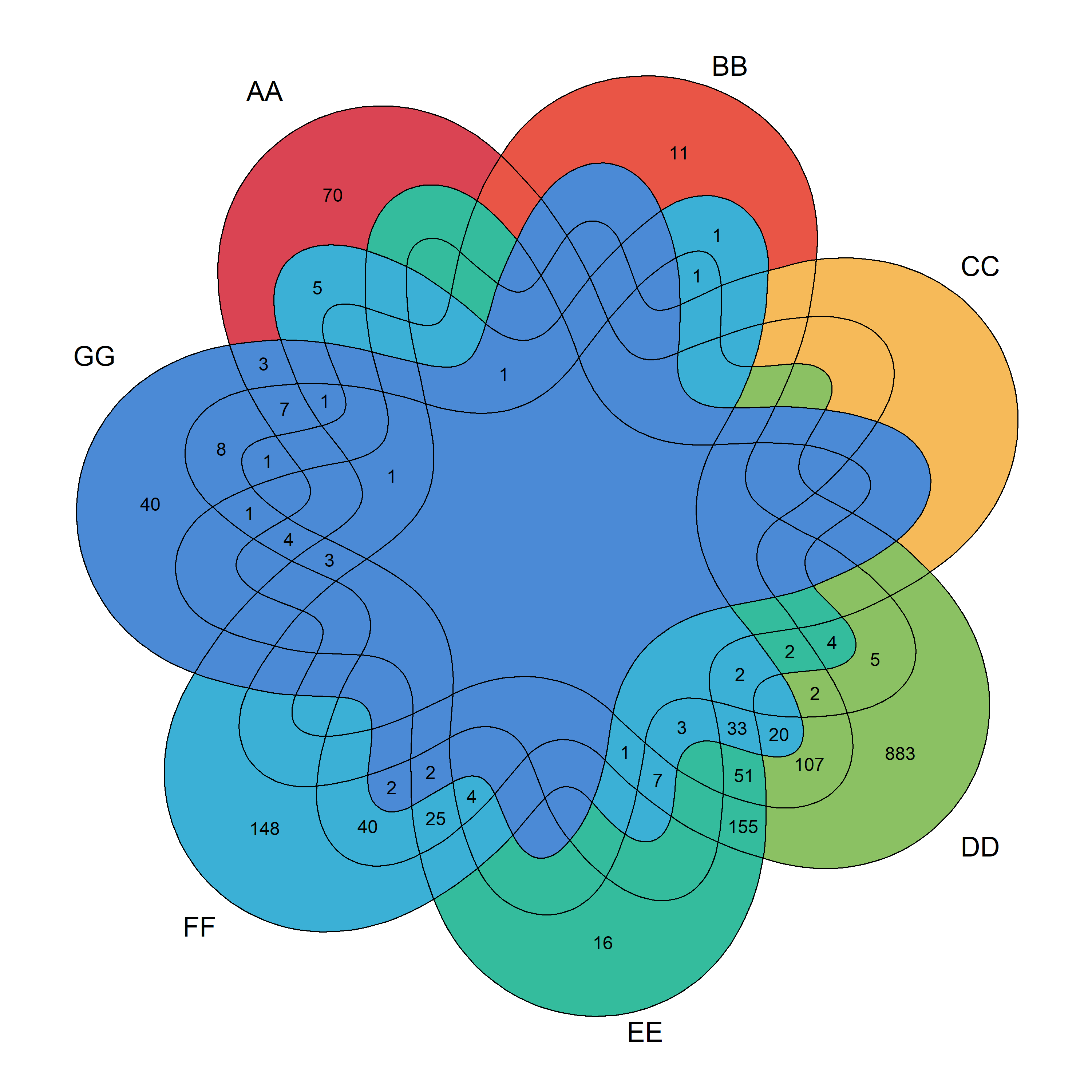
Add module: Add a data visualization module.
-
Version 1.3.0 - January 05, 2024
Add plugins: Developed a local plugin tool (GWASkit) for large files.
-
Version 1.2.0 - February 22, 2024
Add plugins: Complete the development of the plugin module.
-
Version 1.1.0 - December 25, 2023
Analysis Module: Complete the development of the Mendelian analysis module.
-
Version 1.0.0 - December 13, 2023
MRanalysis online, simplifying Mendelian analysis.
Quick Links
Doing MR analysis and visualization without coding expertise.
- All
- MR (API)
- MR (Local)
- Plugins
- Plugins (GWASkit)
- Data Visualization