Step 6. Collect Information About the Current R Session.
Contact Us
The Centre for Artificial Intelligence Driven Drug Discovery (AIDD) at Macao Polytechnic University
GO Enrichment Analysis
Gene Ontology (GO) term enrichment is a technique for interpreting sets of genes making use of the Gene Ontology system of classification, in which genes are assigned to a set of predefined bins depending on their functional characteristics.
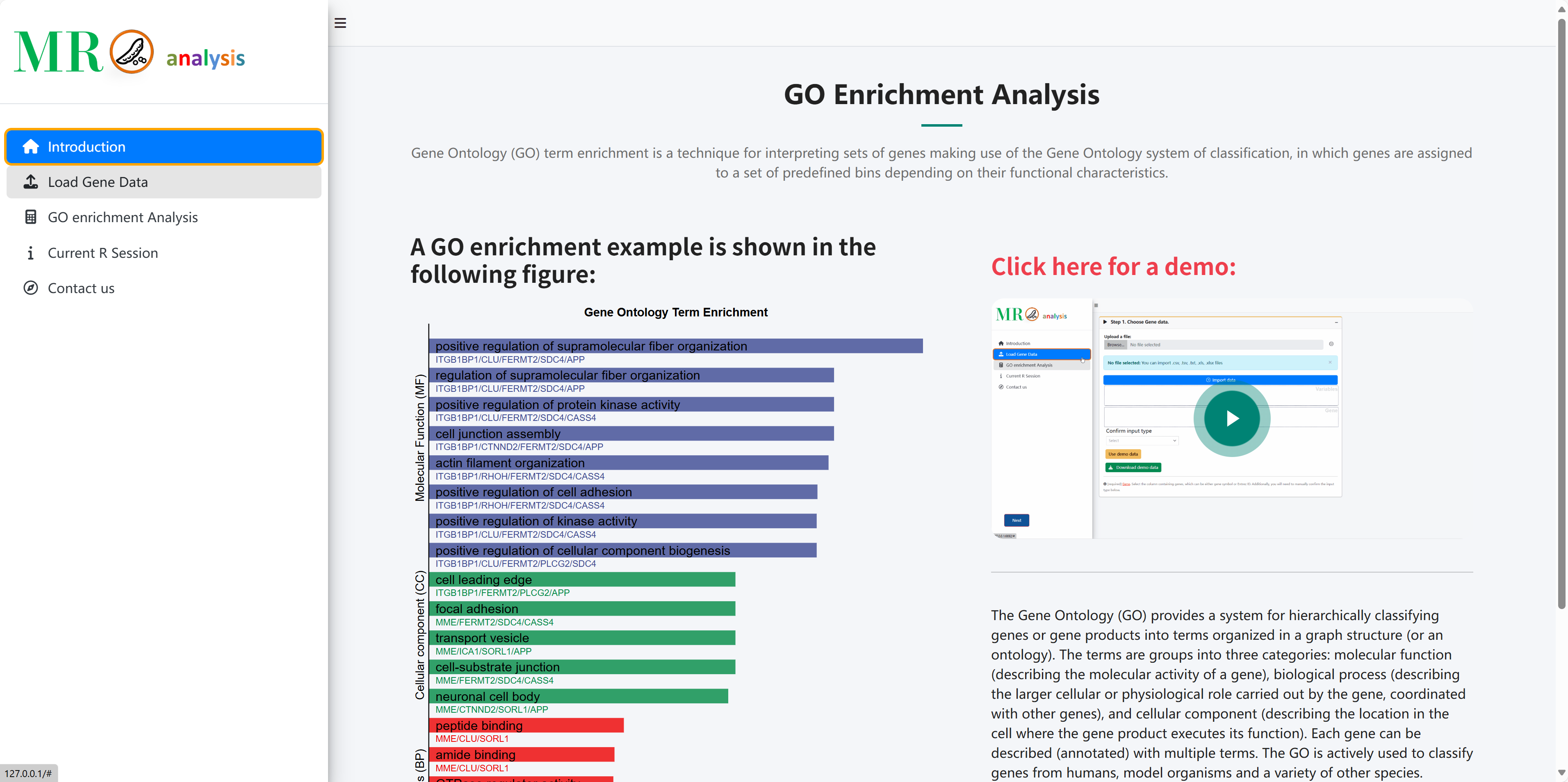
A GO enrichment example is shown in the following figure:
Click here for a demo:
The Gene Ontology (GO) provides a system for hierarchically classifying genes or gene products into terms organized in a graph structure (or an ontology). The terms are groups into three categories: molecular function (describing the molecular activity of a gene), biological process (describing the larger cellular or physiological role carried out by the gene, coordinated with other genes), and cellular component (describing the location in the cell where the gene product executes its function). Each gene can be described (annotated) with multiple terms. The GO is actively used to classify genes from humans, model organisms and a variety of other species.
This application is conducted using clusterProfiler[1] (v4.6.2) in R.
[1] Xu, S., Hu, E., Cai, Y. et al. Using clusterProfiler to characterize multiomics data. Nat Protoc (2024). https://doi.org/10.1038/s41596-024-01020-z
Step 1. Choose Gene data.
[required] Gene. Select the column containing genes, which can be either gene symbol or Entrez ID. Additionally, you will need to manually confirm the input type below.