Step 6. Collect Information About the Current R Session.
Contact Us
The Centre for Artificial Intelligence Driven Drug Discovery (AIDD) at Macao Polytechnic University
SNP Gene Mapping and Enrichment
MAGMA: Generalized Gene-Set Analysis of GWAS Data.
MAGMA is one the most commonly used tools for gene-based and gene-set analysis.
Click here for a demo:
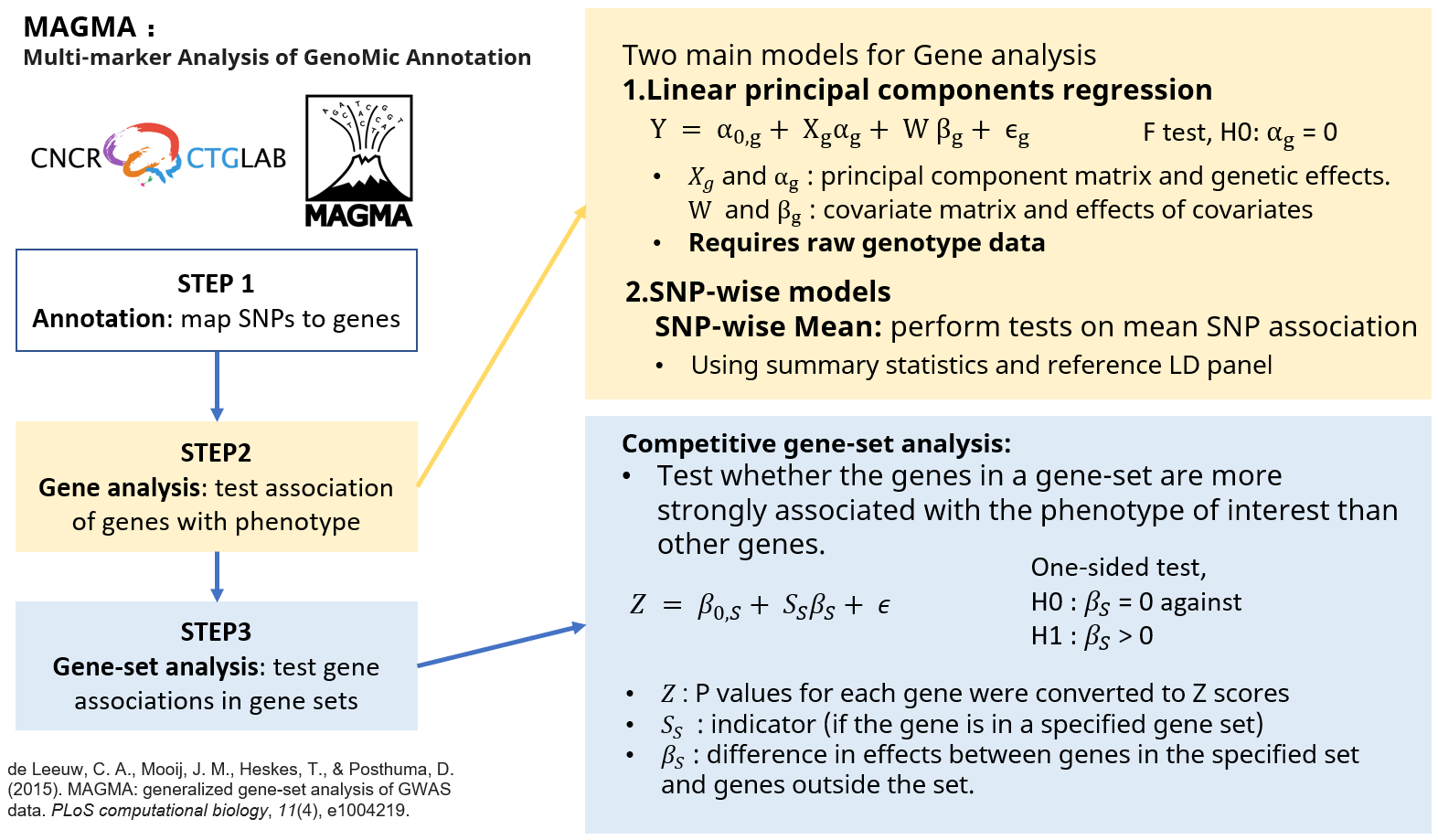
MAGMA Introduction
MAGMA employs a multiple linear principal components regression, and F test to obtain P values for genes.
The workflow for performing MAGMA is as follows (See details):
- Install MAGMA
- Download reference files
- Format input files
- Annotate SNPs
- Gene-based analysis
- Gene-set level analysis
This application is conducted using MAGMA[1] (v1.10).
[1] de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015 Apr 17;11(4):e1004219. doi: 10.1371/journal.pcbi.1004219. PMID: 25885710; PMCID: PMC4401657.
Step 1. Choose SNP data.
[required] SNP, namely SNP ID in dbSNP databases, an it is an identifier for a SNP, which represents a specific variation in the genome.
[required] CHR indicates the chromosome on which the SNP is located in the genome (1, 2, 3, .., X, Y).
[required] POS denotes the position of the SNP on the chromosome.
[optional] P.VALUE is a statistical measure used to assess the assocaition between a SNP and a studied trait, such as a disease.
If this value is NULL, perform gene analysis on raw GWAS data. Otherwise, perform gene analysis on SNP P-values.