Step 6. Collect Information About the Current R Session.
Contact Us
The Centre for Artificial Intelligence Driven Drug Discovery (AIDD) at Macao Polytechnic University
A Universal Enrichment Analyzer
Functional enrichment analysis plays a key role in interpreting high-throughput omics data in life sciences.
Click here for a demo:
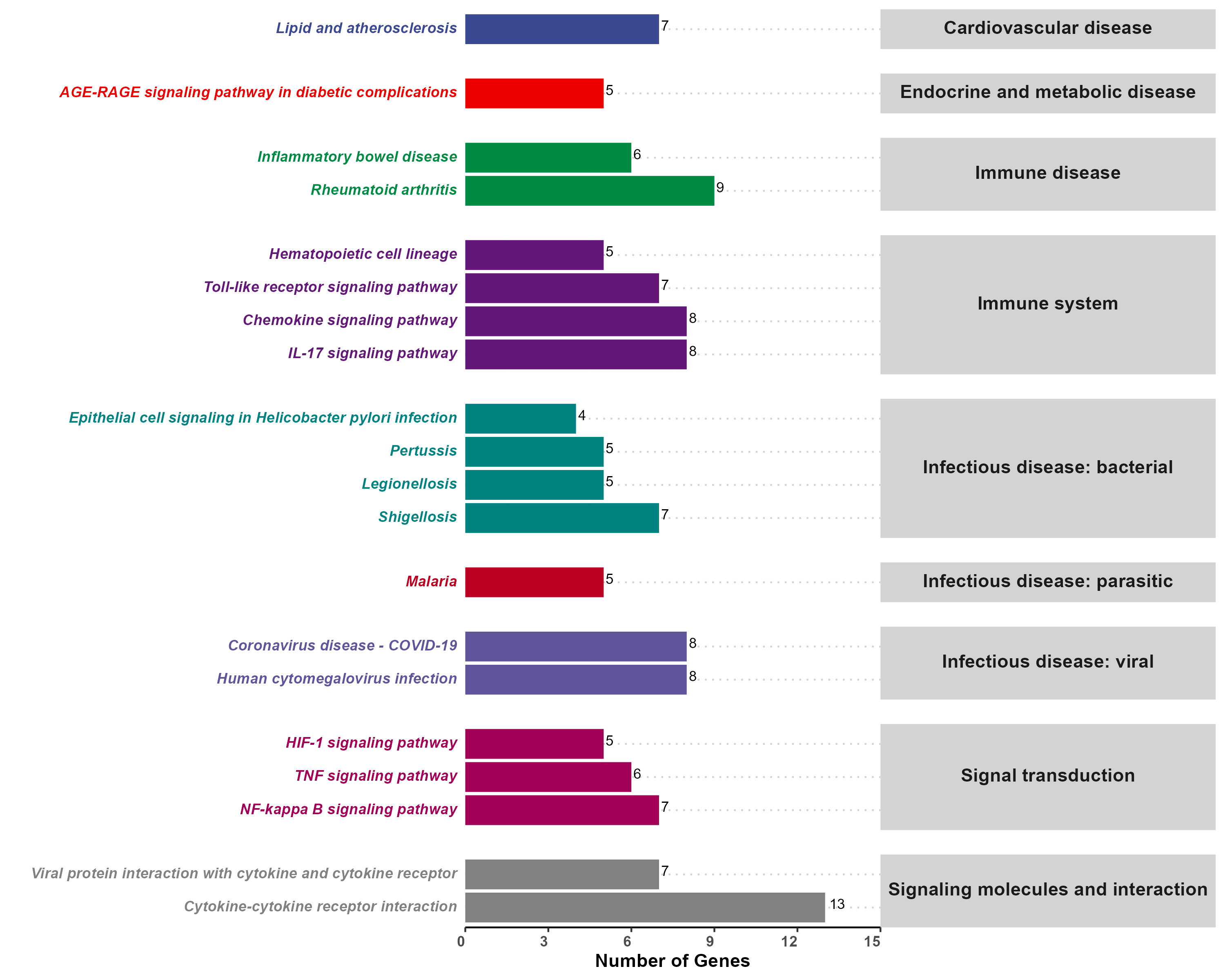
Enrichment analysis is a method used in bioinformatics and computational biology to identify categories (e.g., biological processes, molecular functions, pathways) that are overrepresented or underrepresented in a given set of genes, proteins, or other biological entities. It helps to interpret high-throughput omics data by associating the data with known biological knowledge, such as gene ontology (GO) terms, KEGG pathways, or other functional annotations.
This application is conducted using clusterProfiler[1] (v4.6.2) in R.
[1] Xu, S., Hu, E., Cai, Y. et al. Using clusterProfiler to characterize multiomics data. Nat Protoc (2024). https://doi.org/10.1038/s41596-024-01020-z
Applications:
Understanding biological processes: Enrichment analysis helps in understanding which biological processes or pathways are associated with differentially expressed genes in transcriptomics studies.
Biomarker discovery: Enrichment analysis can aid in identifying potential biomarkers for diseases by associating gene sets with specific functional categories.
Drug discovery: Pathway or GO enrichment analysis can be used to explore the molecular mechanisms of drug action or to identify potential drug targets.
Step 1. Choose Gene/Protein/Metabolite/... data.
[required] pathway, select one column, the id or name of the pathway in the pathway database.
[required] pathway, select one column, the id or name of the pathway in the pathway database.
